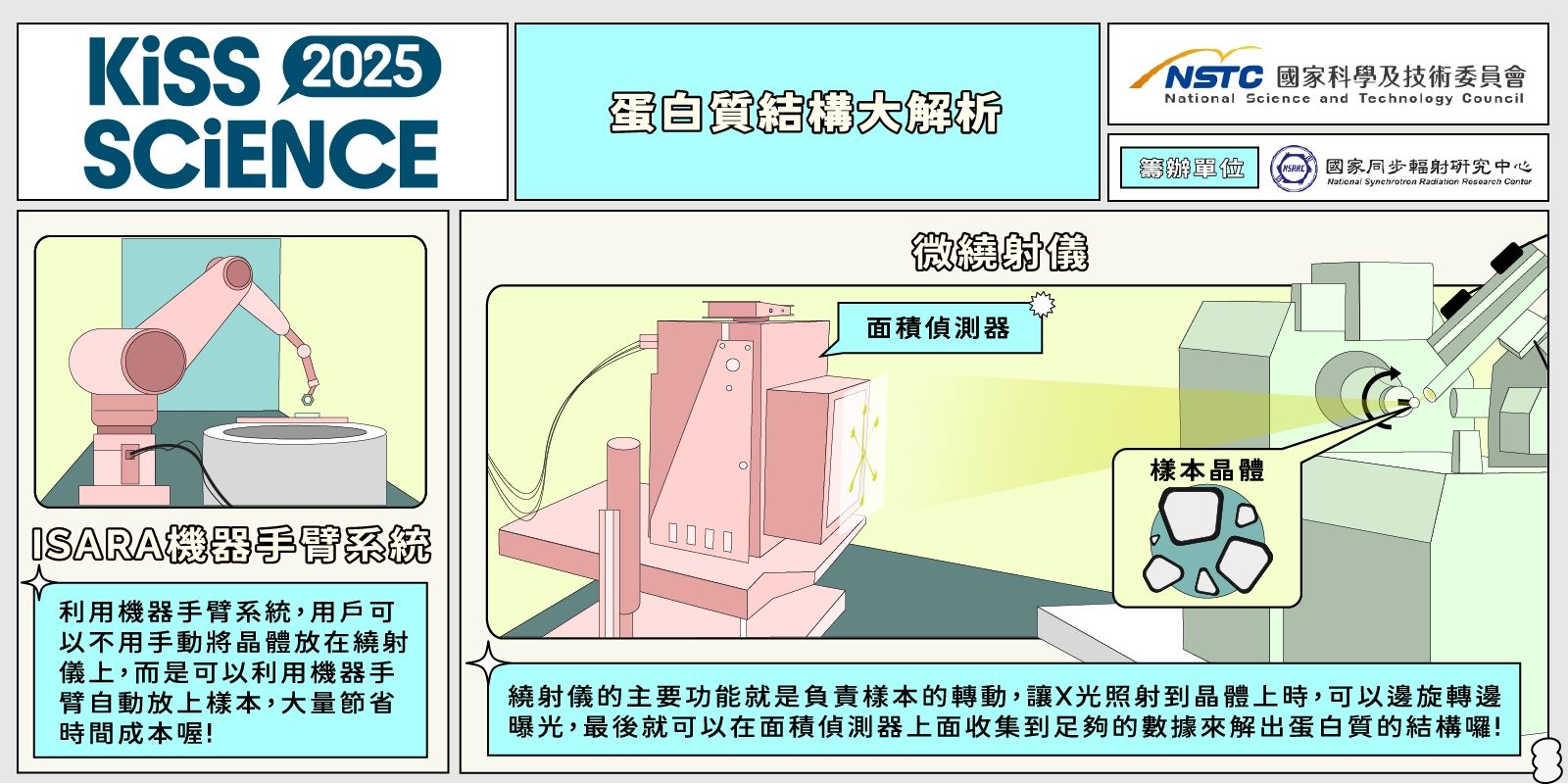
The core technique of our facility is Synchrotron Radiation Protein Crystallography, a technique that uses the principle of single crystal X-ray diffraction to determine high-resolution three-dimensional protein molecular structure from a protein crystal. It is the most powerful and important method to study biological structures. Currently, nine out of every ten biological structures are solved by this technique. Worldwide, only 18 countries and the European Union possess synchrotron radiation facilities and provide protein crystallography technique, and Taiwan is included. Being the only facility in Taiwan that provides this technique, our goal is to build a world-class protein crystallographic facility to assist industrial and academic scientists and experts in the field of biomedical science (including structural biology, proteomics, biochemistry, molecular biology, drug design, biotechnology, etc.) to develop prevention, diagnosis, and treatment of various diseases. That helps improve the health of people, increase the quality of life and effectively conserve medical resources.
| NSRRC PX Beamline |
Micro-focus Protein Crystallography |
Protein Microcrystallography |
Biopharmaceutical Protein Crystallography |
|---|---|---|---|
| X-ray source | TPS/ 3 GeV/ Undulator | TPS/ 3 GeV/ Undulator | TLS/ 1.5 GeV/ Wiggler |
| General user time | 70% | 75% | 80% |
| Status | Operational | Operational | Operational |
| Experiment | MAD/Micro-focus beam | MAD/Microbeam | MAD |
| Energy range (keV) | 6.0-20.0 | 5.7-20.0 | 5.6-15.5 |
| Wavelength (Å) | 2.06-0.62 | 2.18-0.62 | 2.21-0.80 |
| Aperture Size (µm) (types)* | 100-2 | 200-10 (9)* | 200-50 (5)* |
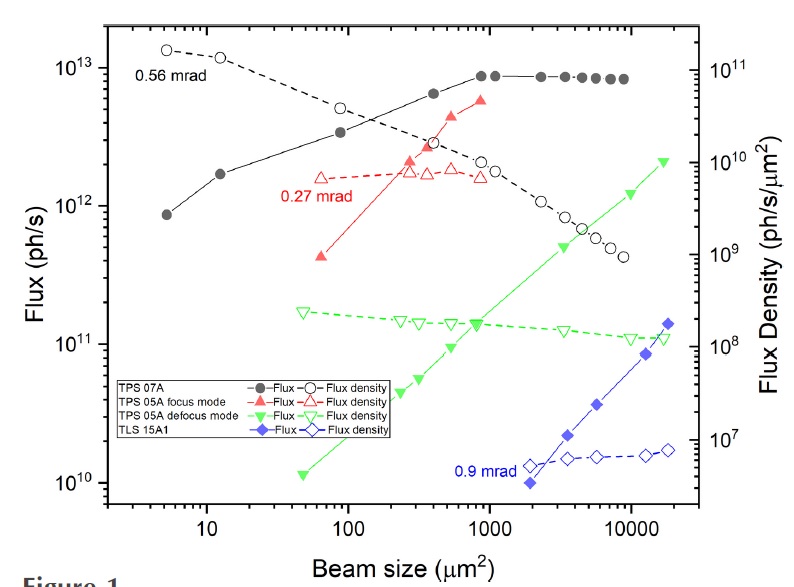
| Flux After Aperture (p/s) | 820-86×1010 | 570-4.0×1010 | 14/8.5/3.7/2.2/1.0×1010 |
| Flux Density (phots/s/µm2) | 9.4-1600×108 | 1.8-5.0×108 | 5.7-7.5×106 |
| Time to Henderson Limit (s) | 0.24-42.6 | 12.1-15.4 | 5333-7018 |
| Detector | EIGER2 X 16M | EIGER2 X 9M | MX300HE |
| Frame Rate (frames/s) | 130 Hz | 230 Hz | 1 |
| Sample Changer | ISARA | ISARA | SAM |
| Remote Access | YES | YES | YES |
- Chung-Kuang Chou, Chien-Chang Tseng, Cheng-Hung Chiang, Yi-Hui Chen, Yi-Chun Liu, Chen-Ying Huang, Chun-Hsiung Chao and Chun-Hsiang Huang, The current status and future prospects of the Synchrotron Radiation Protein Crystallography Core Facility at NSRRC: a focus on the TPS 05A, TPS 07A and TLS 15A1 beamlines. J. Synchrotron Rad. 2025, 32(2), 457. [doi: 10.1107/S1600577525000177]
- Cheng-Hung Chiang, Chung-Kuang Chou, Chien-Chang Tseng, Yi-Hui Chen, Yi-Chun Liu, Chen-Ying Huang, Chun-Hsiung Chao, and Chun-Hsiang Huang, Biopharmaceutical Beamline TLS 15A1 for Macromolecular Crystallography at the National Synchrotron Radiation Research Center. J. Chin. Chem. Soc. 2024, 71(7), 721. [doi: 10.1002/jccs.202400111]
- Yi-Hui Chen, Chien-Chang Tseng, Chung-Kuang Chou, Yi-Chun Liu, Cheng-Hung Chiang, Chen-Ying Huang, Chun-Hsiung Chao and Chun-Hsiang Huang, The highly efficient protein crystallography beamline TLS 13B1 at the National Synchrotron Radiation Research Center. J. Chin. Chem. Soc. 2023, 70(5), 1219. [doi: 10.1002/jccs.202300057]
- Min-Guan Lin et al., Unraveling the structure and function of a novel SegC protein interacting with the SegAB chromosome segregation complex in Archaea. Nucleic Acids Res. 2024, 52(16), 9966.[doi: 10.1093/nar/gkae660]
- Ko-Ting Liu et al., Structural insights into the assembly of type IIA topoisomerase DNA cleavage-religation center. Nucleic Acids Res. 2024, 52(16), 9788. [doi: 10.1093/nar/gkae657]
- Shih-Chun Huang et al., Targeting DNA junction sites by bis-intercalators induces topological changes with potent antitumor effects. Nucleic Acids Res. 2024, 52(15), 9303. [doi: 10.1093/nar/gkae643]
- Shan-Meng Lin, et al., Structural basis of water-mediated cis Watson-Crick/Hoogsteen base-pair formation in non-CpG methylation. Nucleic Acids Res. 2024, 52(14), 8566. [doi: 10.1093/nar/gkae594]
- Chen-Hsi Chu et al., Insights into the molecular mechanism of ParABS system in chromosome partition by HpParA and HpParB. Nucleic Acids Res. 2024, 52(12), 7321. [doi: 10.1093/nar/gkae450]
- Eric Y C Mao, Han-Yi Yen, and Chyuan-Chuan Wu, Structural basis of how MGME1 processes DNA 5' ends to maintain mitochondrial genome integrity. Nucleic Acids Res. 2024, 52(7), 4067. [doi: 10.1093/nar/gkae186]
- Po-Yin Chen, et al., A whole-cell platform for discovering synthetic cell adhesion molecules in bacteria. Nat Commun. 2024, 15(1), 6568. [doi: 10.1038/s41467-024-51017-1]
- Mei-Hui Hou, et al., Structural and functional characterization of cyclic pyrimidine-regulated anti-phage system. Nat Commun. 2024, 15(1), 5634. [doi: 10.1038/s41467-024-49861-2]
- Shen Wang, et al., Structural basis for recruitment of peptidoglycan endopeptidase MepS by lipoprotein NlpI. Nat Commun.2024,15(1) ,5461. [doi: 10.1038/s41467-024-49552-y]
- Wesley Tien Chiang, et al., Structural basis and synergism of ATP and Na+ activation in bacterial K+ uptake system KtrAB. Nat Commun. 2024, 15(1), 3850. [doi: 10.1038/s41467-024-48057-y]
- Chao-Yu Yang, et al., Deciphering DED assembly mechanisms in FADD-procaspase-8-cFLIP complexes regulating apoptosis. Nat Commun. 2024, 15(1), 3791. [doi: 10.1038/s41467-024-47990-2]